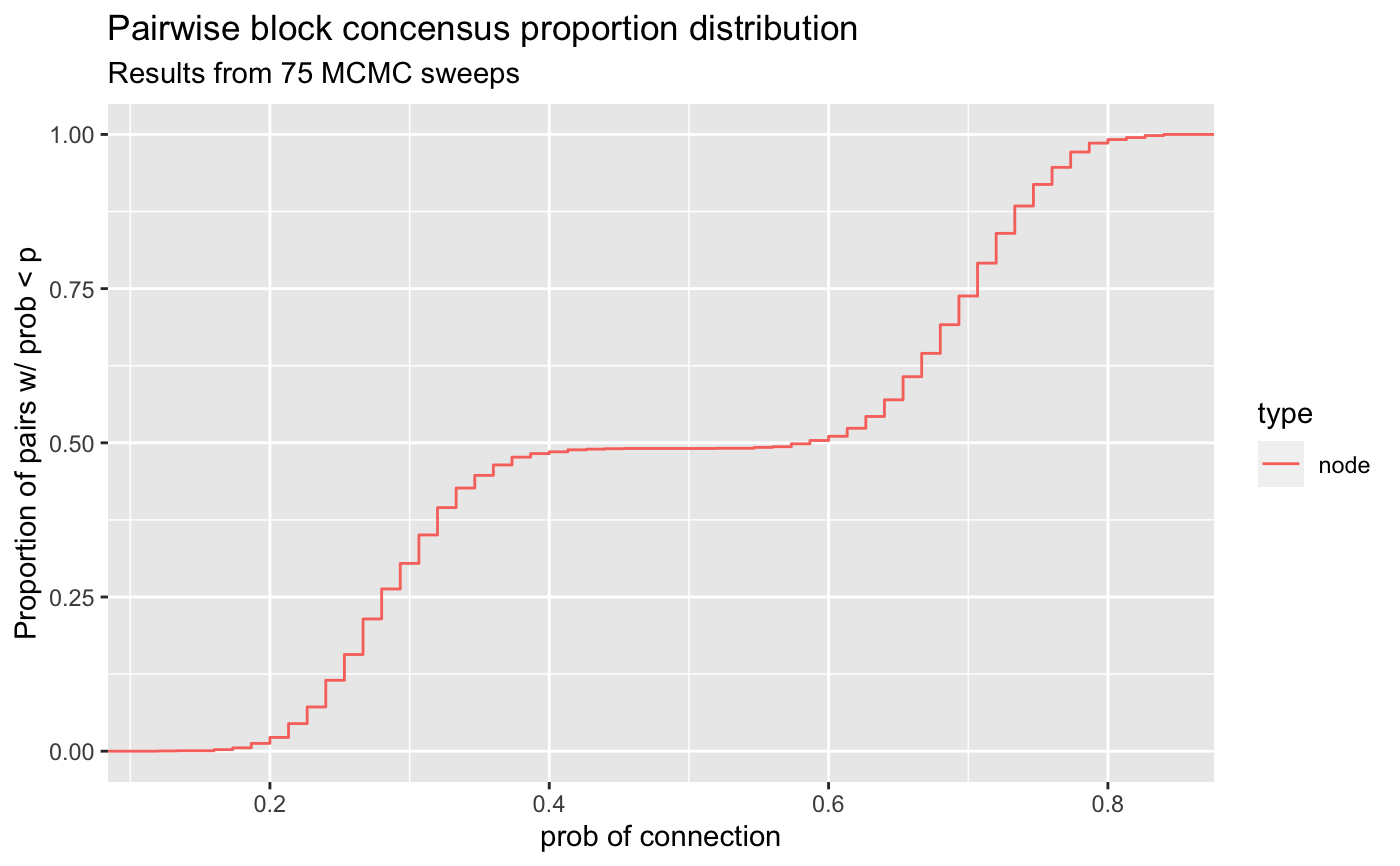
Visualize pairwise propensity distribution
Source:R/visualize_propensity_dist.R
visualize_propensity_dist.RdInvestigate the results of cluster stability from MCMC sweeps. Shows the empirical CDF of the pairwise probability/proportion of two nodes residing in the same block over the given MCMC sweeps.
visualize_propensity_dist(sbm, isolate_type = NULL)
Arguments
| sbm | Object of class |
|---|---|
| isolate_type | Node type to be isolated for visualization. If left empty all types are included. |
Value
GGplot2 plot containing ECDF of the pairwise propensities of residing in same block for MCMC sweeps.
Details
sbmr::mcmc_sweep(sbm, track_pairs = TRUE, ...) must be run on object before this function.
See also
Other visualizations:
visualize_collapse_results(),
visualize_mcmc_trace(),
visualize_network(),
visualize_propensity_network()
Examples
# Simulate a model net <- sim_basic_block_network(n_blocks = 3, n_nodes_per_block = 25) %>% initialize_blocks(3) %>% mcmc_sweep(num_sweeps = 75, track_pairs = TRUE) # Plot results visualize_propensity_dist(net)